bed_intersect: R Equivalent to Bedtools Intersect
bedtools intersect is a useful tool in bioinformatics for finding the overlaps between genomic intervals from the BED file.
bedtools intersect is a part of bedtools utilities which are developed to run as a command-line on a UNIX environment and could not be run on R software.
However, you can use bed_intersect function from the R valr package for performing bedtools intersect analysis in R.
The basic syntax for bed_intersect function is:
# install.packages("valr")
# load package
library("valr")
bed_intersect(file1.bed, file2.bed)
Where, file1.bed and file2.bed contains the genomic intervals in BED format.
The following example explains how to find the overlapping intervals in R using bed_intersect function (similar to bedtools intersect).
Create two example genomic intervals to find the overlap,
# first BED file
bed1 <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 100, 500,
"chr1", 600, 700
)
# second BED file
bed2 <- tibble::tribble(
~chrom, ~start, ~end,
"chr1", 300, 500,
"chr1", 800, 900
)
We have created two genomic intervals in BED format using the tibble. Additionally, you can also import the data from the BED files.
Now, we will perform the bedtools intersect analysis using bed_intersect function to find the overlapping intervals between bed1 and bed2
genomic intervals.
# install.packages("valr")
# load package
library("valr")
bed_intersect(bed1, bed2)
# output
# A tibble: 1 × 6
chrom start.x end.x start.y end.y .overlap
<chr> <dbl> <dbl> <dbl> <dbl> <int>
1 chr1 100 500 300 500 200
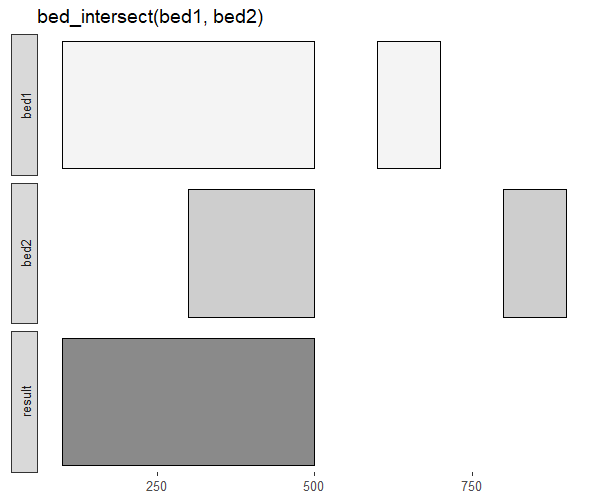
You can see that there is one overlapping interval between the two intervals. There is a 200 bp overlap between the two intervals.
In addition, you can also visualize the overlap between the two genomic intervals using the bed_glyph function.
# load package
library("valr")
bed_glyph(bed_intersect(bed1, bed2))
The additional parameters can also be used with bed_intersect as described here.